About GenDisFinder
Motivation: In this post genomic era, a vast amount of biomedical information from several experimental findings, clinical case report and therapeutics has been stored in text databases such as MEDLINE, PubMed Central, BioMed Central, etc. Proper use of existing domain knowledge from the literature is a prerequisite for any novel research. One of the major tasks in biomedical text mining is the extraction of underlying relation between several genes and disease phenotypes. Gene-disease association data from various high-throughput experiments are hidden in the literature and lack a structured form needed for easy information extraction and visualization. A text mining approach based on gene-disease network analysis could predict the new gene-disease association and also reduce the cost of experiments. Hence there is an urgent need for a text mining with network analysis system that extracts both known and novel gene-disease associations and visualization.
GenDisFinder: Here we introduce a automated new web-based text mining tool named "GenDisFinder" that aids in the extraction of known/novel human gene-disease associations from biomedical literature and further categorize them using networks analysis. GeneDisFinder has different modules and tasks: First task involves gene mention and normalization of gene/protein names using our earlier GM and GN taggers NAGGNER and ProNormz. Second task is the disease mention identification and normalization using OMIM based normalized disease phenotype dictionary. Third task is the identification of semantic relation extraction between genes and diseases using Cartesian product and relation keyword dictionary. Final task is the construction of gene-disease association networks and further network analysis to confirm the extracted associations are categorized as Known, inferred or Novel. We show that our new method is capable of predicting gene-disease association with precision, recall and F-score of 82.84%, 74.07% and 78.20% respectively when evaluated with our GenDisFinder corpus.
Availability and Implementation: GenDisFinder is freely available on the web at http://www.biominingbu.org/GenDisFinder. Website is implemented in Perl, MySQL, Java, Cytoscape web and JavaScript.
| Dr. Suresh Subramani | Prof. Jeyakumar Natarajan |
 |
 |
| PhD sureshsubramani-at-hotmail.com |
Professor n.jeyakumar-at-yahoo.co.in |
GenDisFinder Tutorials
Top
Input Information
The input for GenDisFinder can be a biomedical abstract(s) (plain text/MEDLINE/XML format) Samples inputs are available in the home page. The server provides three different options to upload the input; (i) GenDisFinder Format (ii) MEDLINE Format and (iii) XML Format.
GenDisFinder format (plain text)
Example:
<ID>23143597</ID> <TEXT> The genetic landscape......</TEXT>
<ID>22751098</ID> <TEXT> Activation of the AXL kinase ....</TEXT>
Presence of some special character like Greek letters is not supported by the server.
Top
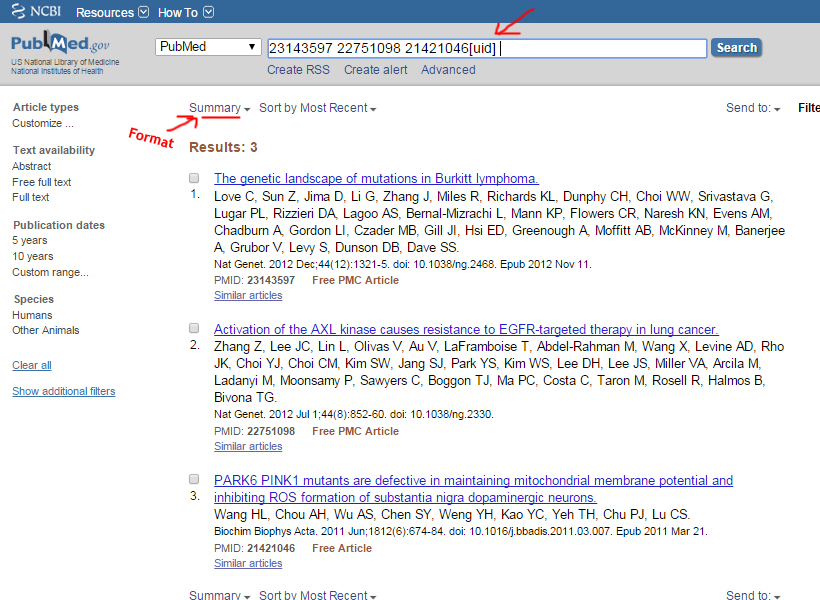
Medline format
Example: http://www.ncbi.nlm.nih.gov/pubmed
List of Pubmed IDs
23143597
22751098
21421046
Keyword search (Any Human OMIM based disease phenotype/Gene name)
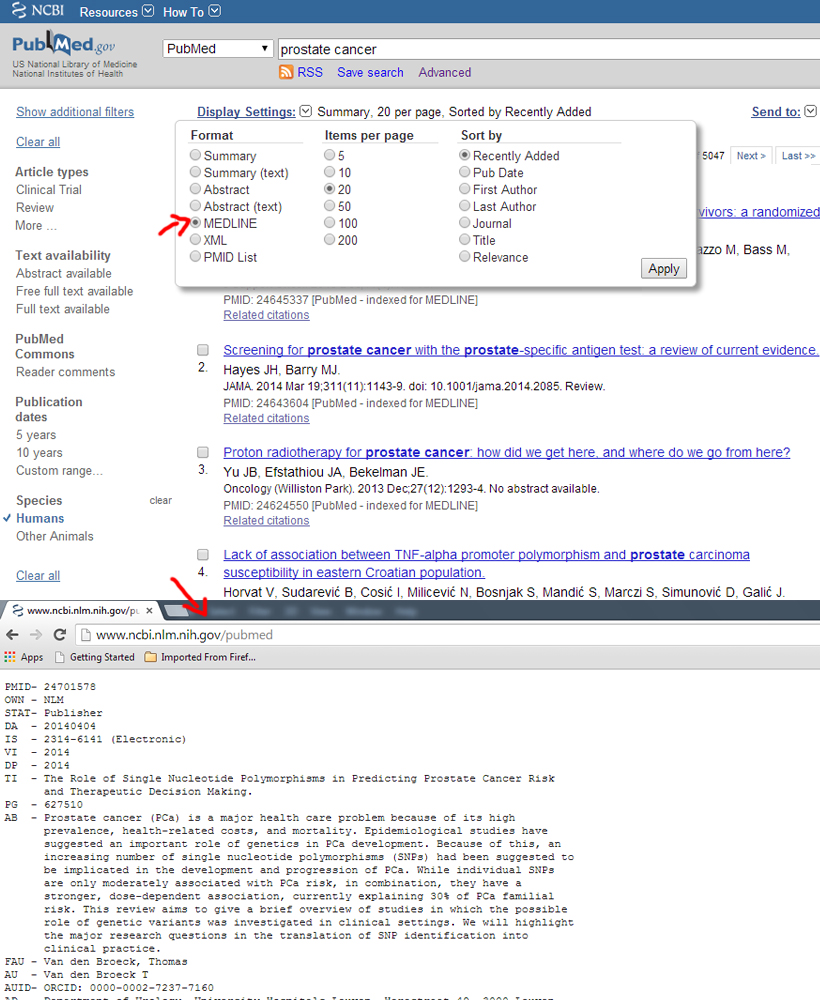
The MEDLINE formats are recommended to be in the NCBI order as this differs between the databases.
Top
XML format
Example: http://www.ncbi.nlm.nih.gov/pubmed
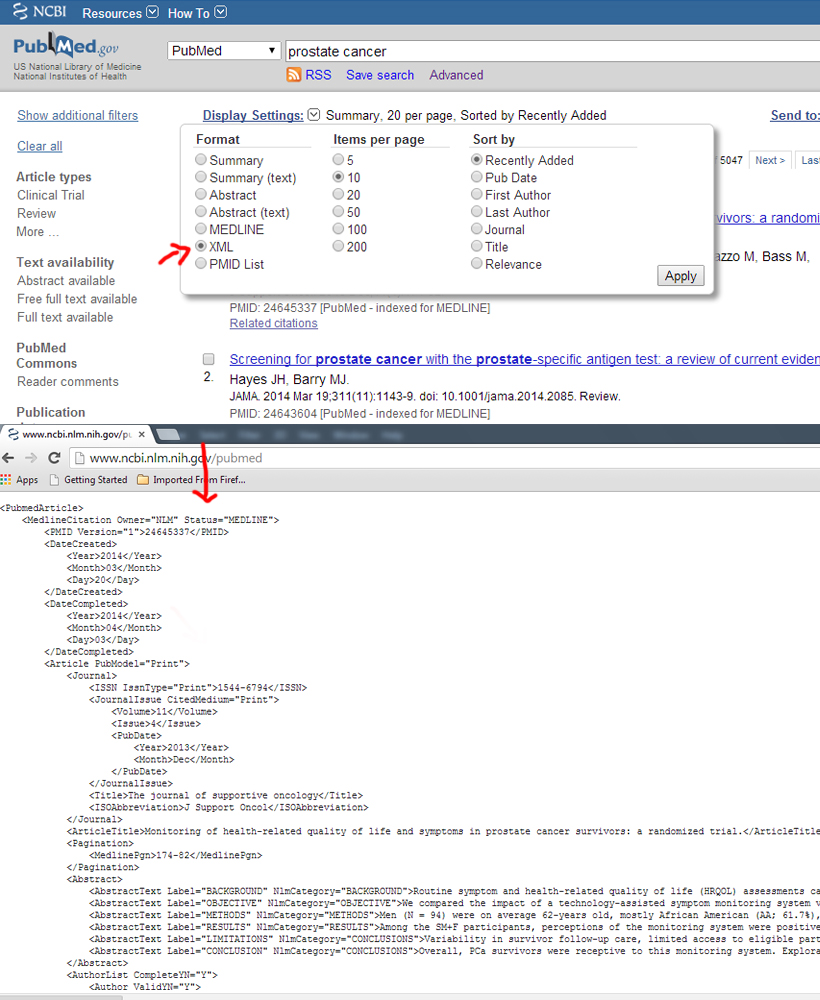
---------------------------------------------------------------------
Top
Output information
Mined Gene-Disease associations from literature
Semantic relation extraction between genes and diseases
Top
Result information
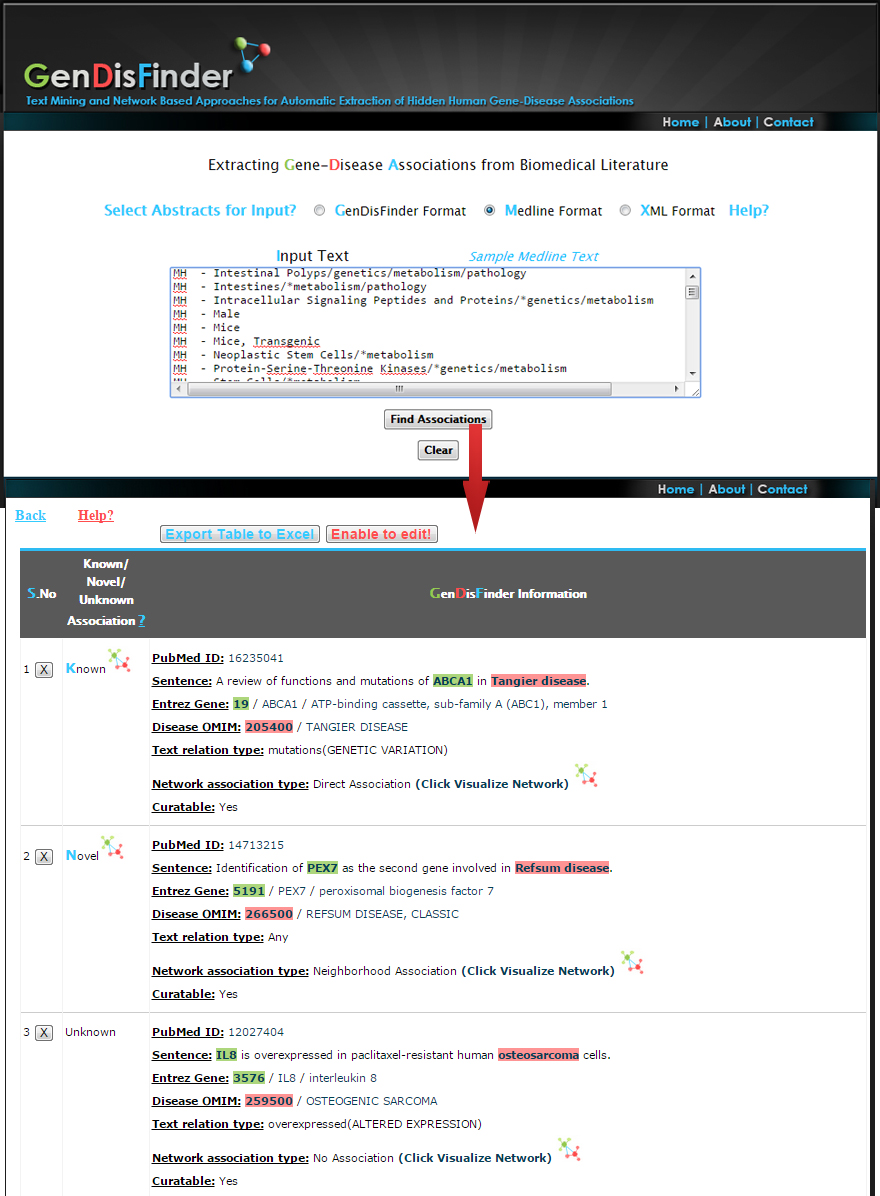
PubMed ID ?: Evidence Pubmed ID
Sentence ?: Evidence Sentence- Red(Disease entity Highlighted) Green(Gene entity Highlighted)
Entrez Gene ?:Normalized Official EntrezGene ID/Official Symbol/Official Full Name
Disease OMIM?:Normalized Disease Phenotype OMIM ID/ Disease Phenotype Name
Text relation type ?:
The relation dictionary is based on the molecular function, where a gene is known to be related with a specific disease. Relation types such as genetic variation, altered expression, regulatory modification, negative association or unrelated relations were recognized as described by Bundschus et al., (2008).
Altered expression (e.g. down-regulated, co-expressed, over-expressed, activated..etc).
Genetic variation (e.g. gene alteration, heritable, mutated, duplication..etc).
Regulatory modification (e.g. methylated, phosphorylation, hypermethylation..etc).
Unrelated (e.g. not, but not, lack of, unrelated, neither..etc).
Any.
Top
Network association type ?:
Diseases with similar phenotypes are often known to share a common set of genes that are functionally related (Brunner and van Driel, 2004). Based on this observation, disease networks are constructed where two diseases are connected, if they share one or more common genes (Goh et al., 2007). The most challenging task is the determination of disease-related genes by performing laborious experiments. A text mining approach based on gene-disease network analysis could predict the gene-disease association and also reduce the cost of such experiments. For example, the extrication of disease genes in molecular networks shows that the genes associated with the same or similar diseases are known to reside in the same neighborhood of these networks and forms functional relations (Oti and Brunner, 2007; Feldman et al., 2008). Genes and diseases are known to be related and hence require a network based framework for the discovery of novel disease genes (Pujana et al., 2007).
This tool aims to discover novel associations between genes and diseases based on direct/neighborhood associations in the network.
Next, for each extracted gene-disease association from literature, we created three human heterogeneous networks namely,
i) Human Disease-Disease similarities (D2D) network was constructed from MimMiner (http://www.cmbi.ru.nl/MimMiner/). The MimMiner contains 5080 x 5080 OMIM disease phenotypes and their similarity information with similarity score derived from the literature (Van Driel et al., 2006)., we used D2D similarities with the similarity score in the range [0.5,1] to include only the highly similar diseases.
ii) Protein-Protein interactions (PPI) network was constructed from Human Protein Reference Database (HPRD) which contains 39376 curated protein-protein interactions (Keshava Prasad et al., 2009)., In the network, the value '1' denotes that the interaction exists and '0' denotes that the interaction does not exist.
iii) Gene-Disease associations (G2D) network was constructed using the OMIM morbid map (Hamosh et al., 2005) and contains 6348 G2D associations. If the gene is known to be associated with the disease, the association score in the matrix is set to be '1'. Otherwise, '0' denotes that the association does not exist.
Then we performed the integrated network analysis of the above three networks using state-of-the-art network association method described by (Guo et al., 2011) and classified extracted gene-disease associations as Known, Novel or Unknown.
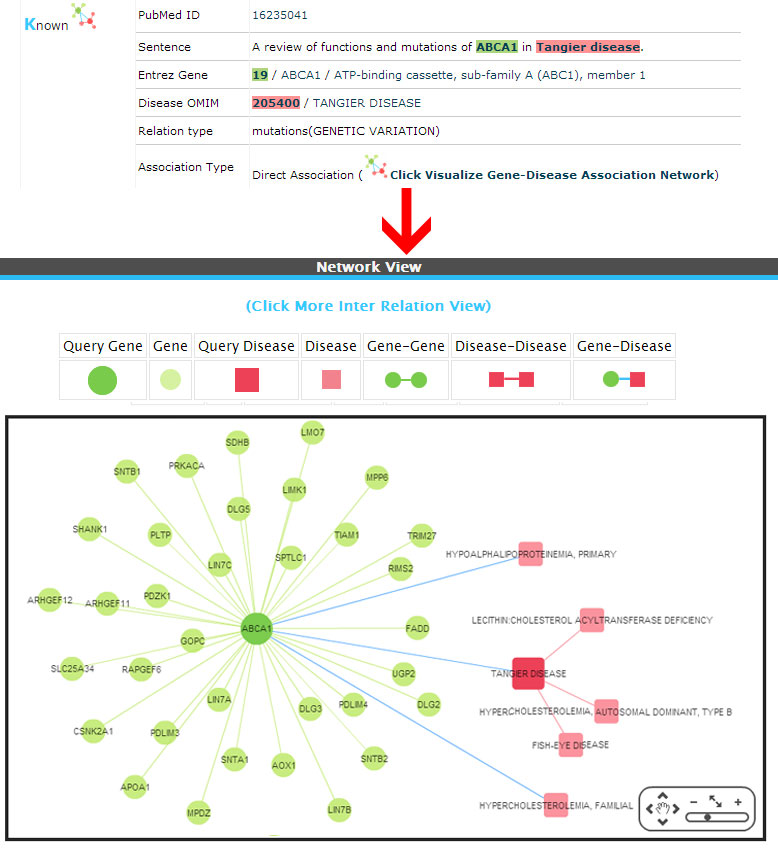
(1) Known (Already a direct association exists between the gene-disease pairs based on the information from databases).
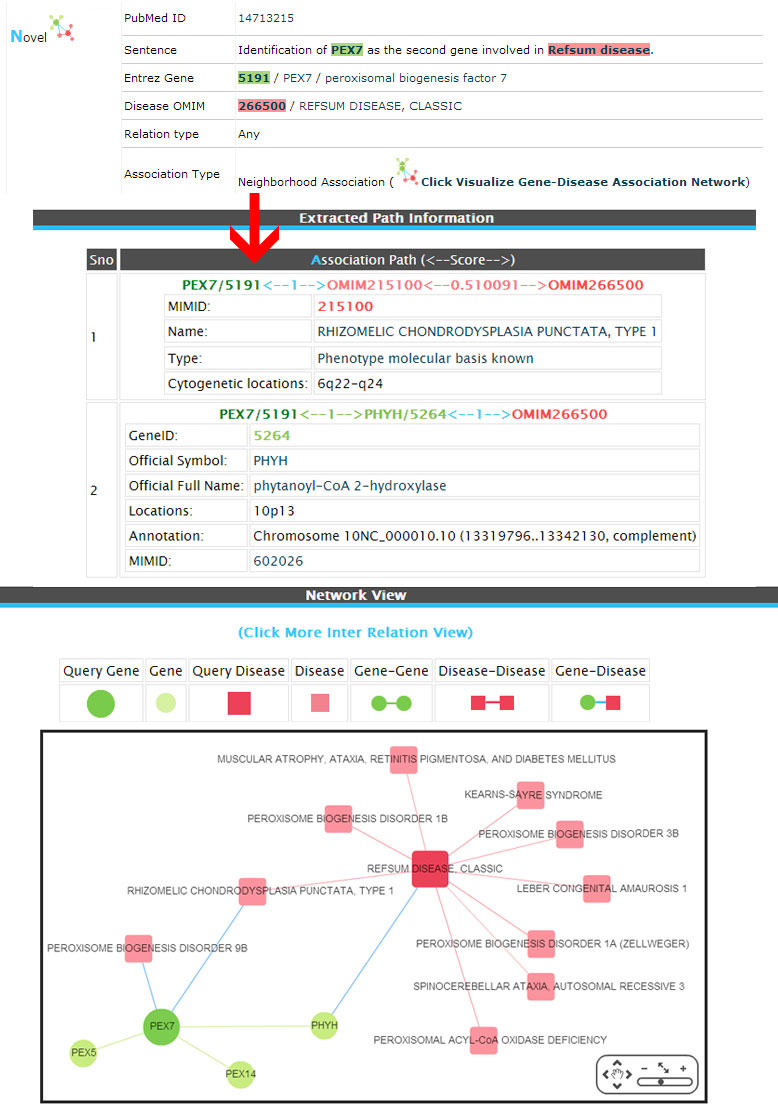
(2) Inferred (The associations are not exists in databases but inferred by network analysis of first neighborhood association between genes/diseases and newly retrieved from the literature.).
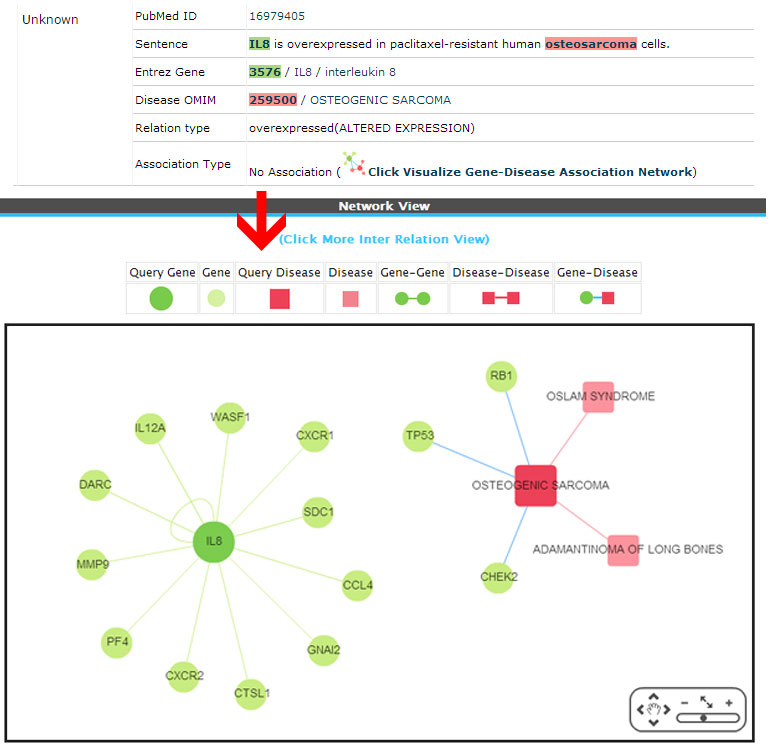
(3) Novel (There is no direct or inferred association from the network. These associations are newly retrieved from the literature).
Top
Known (Direct association)
Example:
Top
Novel (Neighborhood association)
Example:
Top
Unknown ( No association)
Example:
Top
Disclaimer
The GenDisFinder web-based text mining and data mining tool is implemented for academic use only.